Science Consulting - Modelling Projects
Are you looking for ways to simulate your data or model a particular process? Do you want to solve a set of differential equations or are you simply trying to speed up your algorithm?

As part of the many types of consulting services we provide at XpertScientific, we can develop numerical models, carry out simulations, and assist you with finding efficient solutions to your scientific problems. The scientists at XpertScientific have a combined modelling experience of 150+ years and our areas of expertise include include natural and engineering sciences, life and medical sciences, as well as environmental and social sciences.
Recent projects
Some of our recent modelling projects include:
- Monte Carlo simulations and Random Walks in Biology
- Optical ray tracing through free falling rain drops
- Photosynthesis (incl. photo-acclimation & -inhibition) modelling in phytoplankton
- Ocean modelling: 3D at 1/4 degree resolution, using realistic bathymetry and external forcing/data assimilation
- Particle tracking models, both in 1D and 3D
- Turbulence modelling
- Parasite infectivity modelling
- Phylogeny reconstruction, including large scale and genomic datasets. Prediction of optimal phylogenetic markers and barcoding. Molecular dating and ancestral state reconstruction.
- Structural genomics: analysis of the evolution of the genomic content (e.g., transposable elements, repeats, genes) and organisation (e.g., study of gene and genomic region rearrangements).
- Optimization / problem minimization algorithms (least squares, inversion)
Particle tracking in a coastal embayment
The evolution of two different particle clouds in a shallow Mediterranean estuary where the main forcing comes from wind and freshwater inflow, using the Random Walk method from Ross and Sharples (2004).
Kinematic simulation (2D) of turbulence
2D Kinematic Simulation of turbulence based on Fung and Vassilicos (2003). Differently coloured particles are used to visualize the turbulence mixing.
Fraunhofer diffraction at a circular aperture
Electric field amplitude at the detector created by Fraunhofer diffraction at a circular aperture (solved using Runga-Kutta methods).
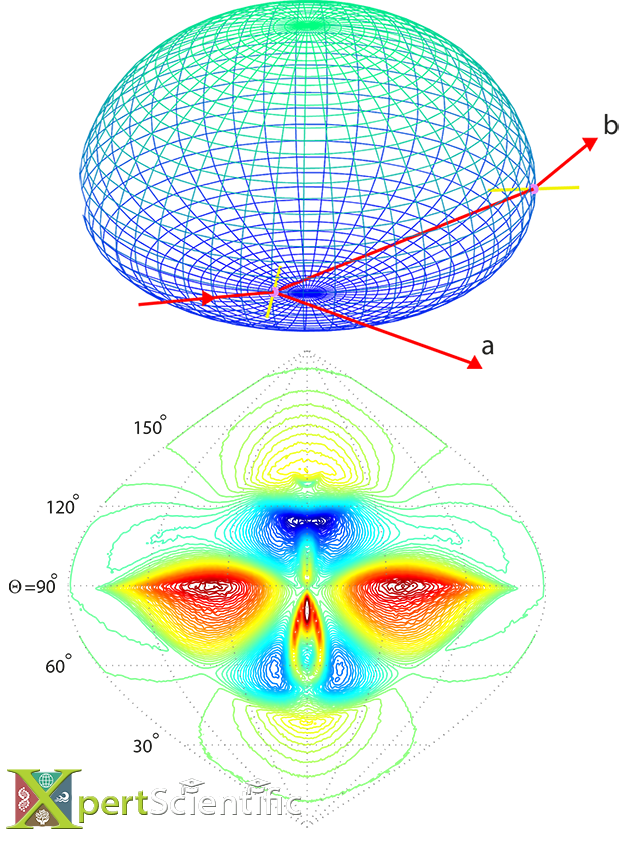
Optical Ray Tracing
Optical ray tracing through free falling rain drops. The top panel shows the ray trace through a 3mm drop with the incoming ray from the left, the surface normals at the entry and exit points (yellow), and the reflected (a) and the twice-refracted forward scattered (b) rays. The bottom panel shows the difference between the y- and z-polarized light intensities of the refraction pattern in the forward hemisphere for a 3mm drop size, using the Monte Carlo model from Ross and Bradley (2002).
Modelling gene order loss
Modelling gene order loss (GOL) from the phylogenetic information of mitochondrial genes in a large group of fungi. The gene order in each species is represented by a set of coloured boxes. GOL can be modelled, based on the distances between species (given by branch lengths), but also from the topology of the tree.
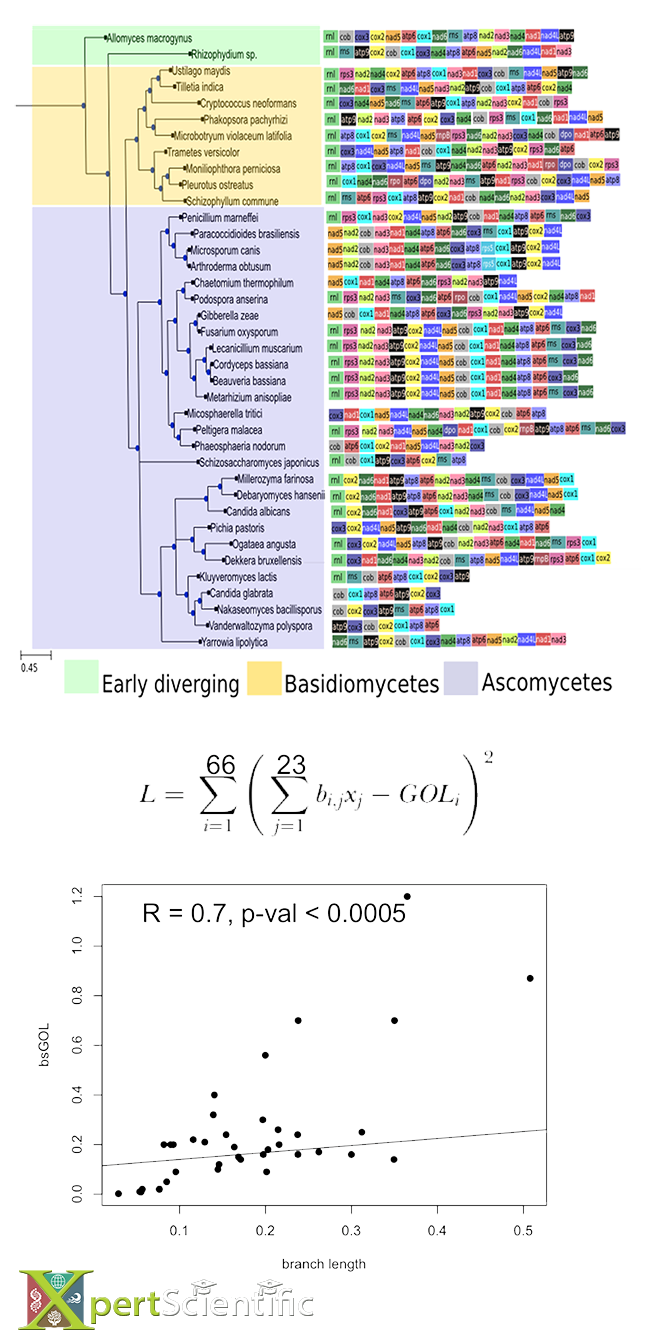
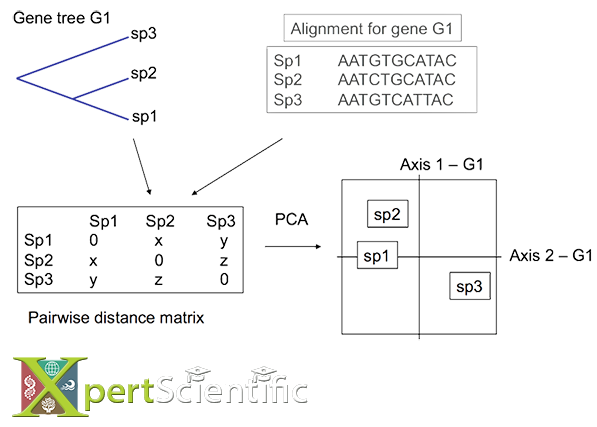
Multivariate analysis
A graphical multivariate approach to detect concordant and discordant (outlier) species in a topology. We extract pairwise distances between species from the tree, perform a principal component analysis, and project the separation axes onto a single plane for a fast visualization.
Programming Languages
We use the following programming languages.
- C/C++
- Fortran
- Matlab
- Python
- R
Some of our clients